tl;dr Although it will be published after StoneAnno, this shape analysis is my first completed research project and technically my first first-authorship, published in Human Brain Mapping. I wrote code in R and MATLAB to fit LMMs to the cortical data from T1w MRI of HD patients and then performed statistical analyses on the results using SurfStat and random field theory. We found that, with a novel method for measuring gyrification, LGI uniquely detects changes in the insula. Of note, I learned that complicated statistical anlayses are uniquely challenging, that I love LMMs and RFT, and that they are too esoteric in the current day – let’s make them more accessible!
Citation
Stoebner, Zachary A., et al. “Comprehensive shape analysis of the cortex in Huntington’s disease.” Human Brain Mapping (2022).
https://doi.org/10.1002/hbm.26125
Background
Abstract
The striatum has traditionally been the focus of Huntington’s disease research due to the primary insult to this region and its central role in motor symptoms. Beyond the striatum, evidence of cortical alterations caused by Huntington’s disease has surfaced. However, findings are not coherent between studies which have used cortical thickness for Huntington’s disease since it is the well-established cortical metric of interest in other diseases. In this study, we propose a more comprehensive approach to cortical morphology in Huntington’s disease using cortical thickness, sulcal depth, and local gyrification index. Our results show consistency with prior findings in cortical thickness, including its limitations. Our comparison between cortical thickness and local gyrification index underscores the complementary nature of these two measures – cortical thickness detects changes in the sensorimotor and posterior areas while local gyrification index identifies insular differences. Since local gyrification index and cortical thickness measures detect changes in different regions, the two used in tandem could provide a clinically relevant measure of disease progression. Our findings suggest that differences in insular regions may correspond to earlier neurodegeneration and may provide a complementary cortical measure for detection of subtle early cortical changes due to Huntington’s disease.
Results
CT
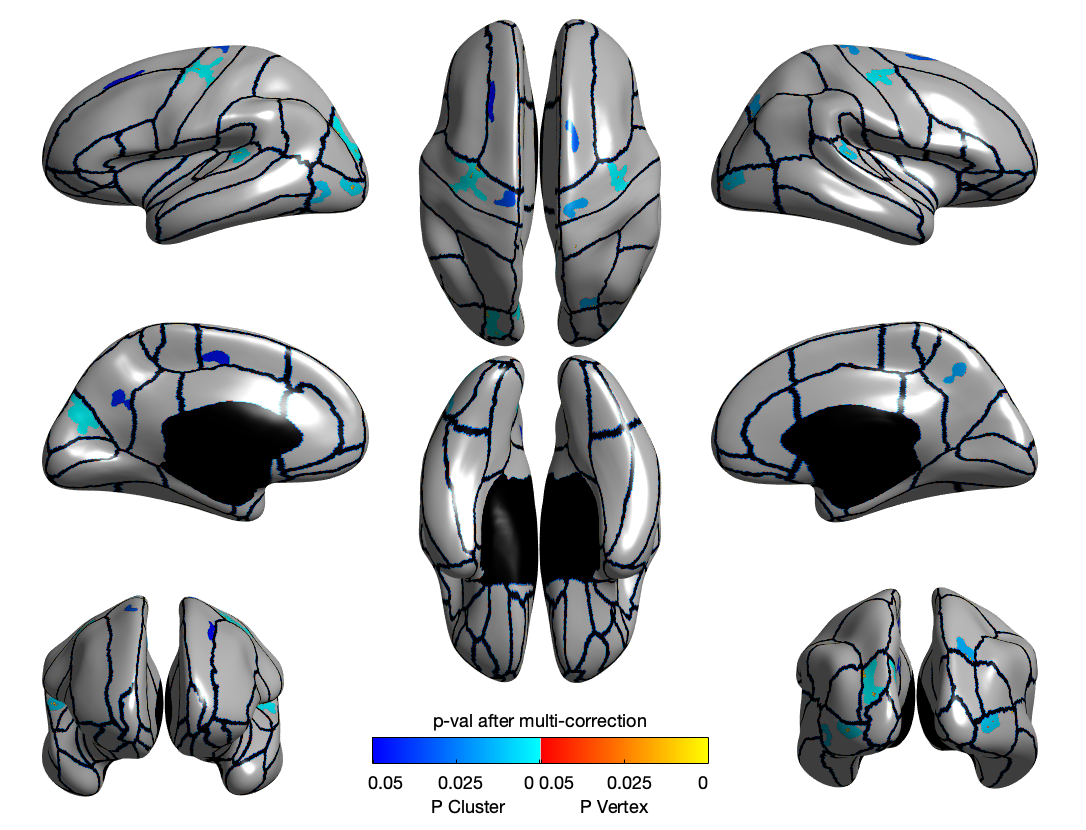
SD
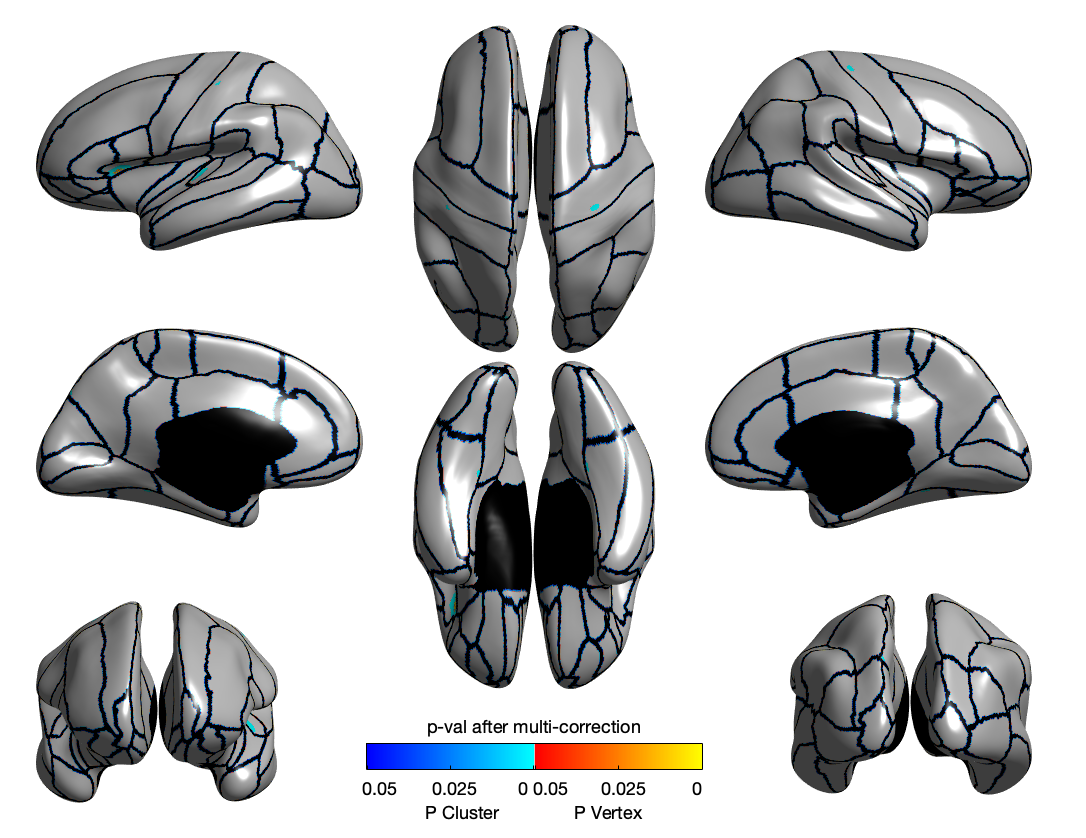
LGI
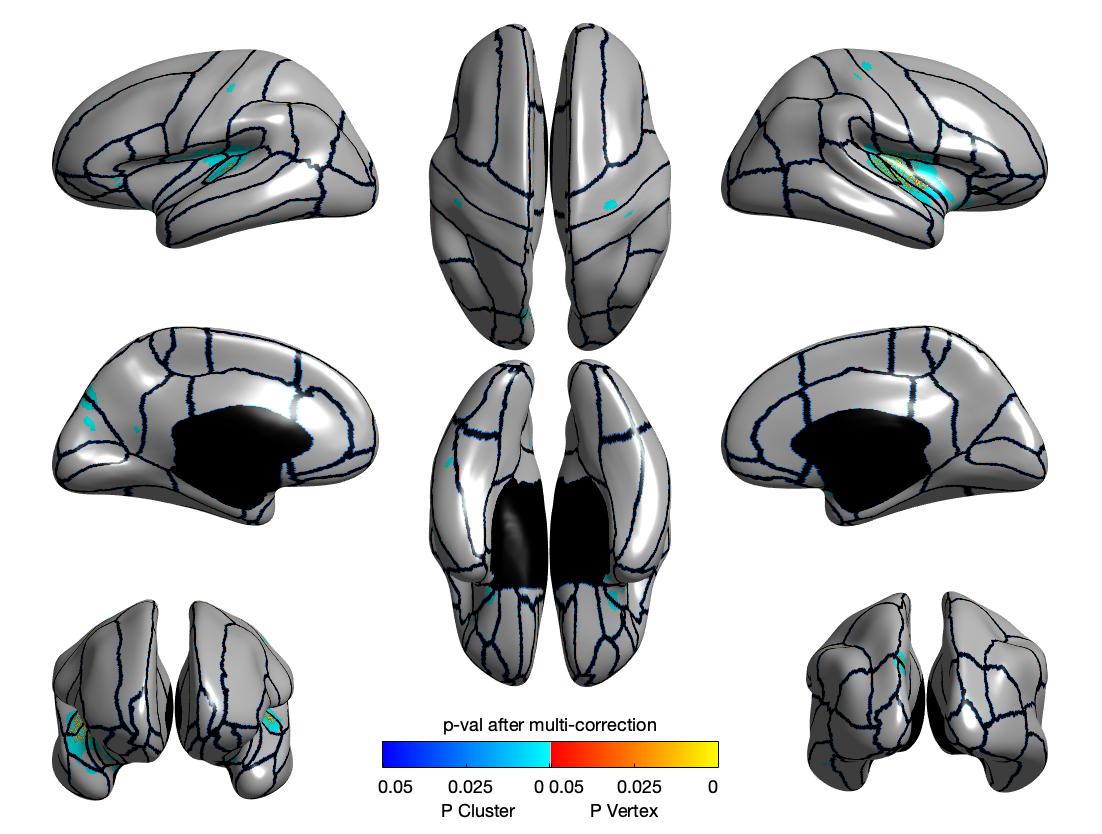
Summary
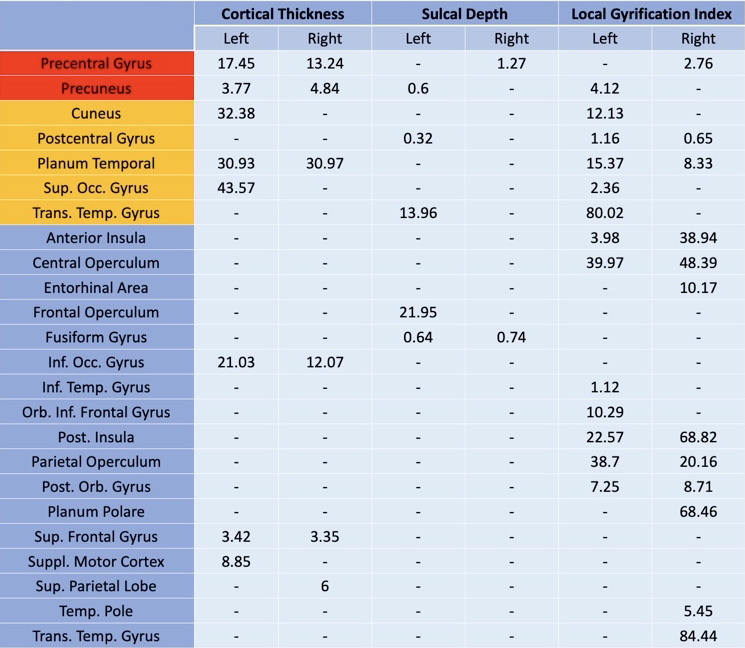
Takeaways
A main takeaway was learning the scientific process in action and, most importantly, learning to work with more experienced researchers. I wrote the code and performed all of the analysis that produced our results. However, I did not develop the awesome acquisition method that generated our LGI data nor the statistical theory behind the analysis. Throughout the project, I have relied heavily on the expertise of my co-authors – all of whom have PhDs whereas I was an undergrad until recently. This first journey in research has been inspiring and indelible. I am beyond grateful for it!
References
-
Walker FO. Huntington’s disease. Lancet. 2007;369(9557):218-228. doi:10.1016/S0140-6736(07)60111-1
-
Long JD, Lee JM, Aylward EH, et al. Genetic Modification of Huntington Disease Acts Early in the Prediagnosis Phase. Am J Hum Genet. 2018;103(3):349-357. doi:10.1016/j.ajhg.2018.07.017
-
Paulsen JS, Ph D, Long JD, et al. Prediction of manifest Huntington disease with clinical and imaging measures : A 12-year prospective observational study. 2015;13(12):1193-1201. doi:10.1016/S1474-4422(14)70238-8.Prediction
-
Ehrlich ME. Huntington’s Disease and the Striatal Medium Spiny Neuron: Cell- Autonomous and Non-Cell-Autonomous Mechanisms of Disease. Neurotherapeutics. 2012;9(2):270-284. doi:10.1007/s13311-012-0112-2
-
Hett K, Johnson H, Coupe P, Paulsen JS, Long JD, Oguz I. Tensor-Based Grading: A Novel Patch-Based Grading Approach for the Analysis of Deformation Fields in Huntington’s Disease. Proc - Int Symp Biomed Imaging. 2020;2020-April:1091-1095. doi:10.1109/ISBI45749.2020.9098692
-
Li H, Zhang H, Johnson H, Long J, Paulsen J, Oguz I. Longitudinal subcortical segmentation with deep learning. In: SPIE Medical Imaging 2021: Image Processing. International Society for Optics and Photonics; 2021:115960D. doi:https://doi.org/10.1117/12.2582340
-
Li H, Zhang H, Hu D, et al. Generalizing MRI subcortical segmentation to Neurodegeneration. In: MLCN Workshop, MICCAI. Springer, Cham; 2020:139-147. doi:https://doi.org/10.1007/978-3-030-66843-3_14
-
Li H, Zhang H, Johnson H, Long J, Paulsen J, Oguz I. MRI Subcortical Segmentation In Neurodegeneration with Cascaded 3D CNNs. In: SPIE Medical Imaging 2021: Image Processing. International Society for Optics and Photonics; 2021:115960W. doi:https://doi.org/10.1117/12.2582005
-
Paulsen JS, Nopoulos PC, Aylward E, et al. Striatal and white matter predictors of estimated diagnosis for Huntington disease. Brain Res Bull. 2010;82(3-4):201-207. doi:10.1016/j.brainresbull.2010.04.003
-
Hedreen JC, Peyser CE, Folstein SE, Ross CA. Neuronal loss in layers V and VI of cerebral cortex in Huntington’s disease. Neurosci Lett. 1991;133(2):257-261. doi:10.1016/0304-3940(91)90583-F
-
Rosas HD, Liu AK, Hersch S, et al. Regional and progressive thinning of the cortical ribbon in Huntington’s disease. Neurology. 2002;58(5):695-701. doi:10.1212/WNL.58.5.695
-
Nopoulos PC, Aylward EH, Ross CA, et al. Cerebral cortex structure in prodromal Huntington disease. Neurobiol Dis. 2010;40(3):544-554. doi:10.1016/j.nbd.2010.07.014
-
Tabrizi SJ, Langbehn DR, Leavitt BR, et al. Biological and clinical manifestations of Huntington’s disease in the longitudinal TRACK-HD study: cross-sectional analysis of baseline data Sarah. 2013;8(9):791-801. doi:10.1016/S1474-4422(09)70170- X.Biological
-
Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A. 2000;97(20):11050-11055. doi:10.1073/pnas.200033797
-
Lyu I, Kang H, Woodward ND, Landman BA. Sulcal depth-based cortical shape analysis in normal healthy control and schizophrenia groups. 2018;1057402(March 2018):1. doi:10.1117/12.2293275
-
Lyu I, Kim SH, Girault JB, Gilmore JH, Styner MA. A cortical shape-adaptive approach to local gyrification index. Med Image Anal. 2018;48:244-258. doi:10.1016/j.media.2018.06.009
-
Wu D, Faria A V., Younes L, Ross CA, Mori S, Miller MI. Whole-brain segmentation and change-point analysis of anatomical brain mri—application in premanifest huntington’s disease. J Vis Exp. 2018;2018(136):1-9. doi:10.3791/57256
-
Tan X, Ross CA, Miller MI, Tang X. CHANGEPOINT ANALYSIS OF PUTAMEN AND THALAMUS SUBREGIONS IN PREMANIFEST HUNTINGTON’S DISEASE. In: 2018 IEEE 15th International Symposium on Biomedical Imaging. ; 2018:531-535. doi:10.1109/ISBI.2018.8363632
-
Tang X, Ross CA, Johnson H, et al. Regional subcortical shape analysis in premanifest Huntington’s disease. Hum Brain Mapp. 2019;40(5):1419-1433. doi:10.1002/hbm.24456
-
Hong Y, O’Donnell LJ, Savadjiev P, et al. Genetic load determines atrophy in hand cortico-striatal pathways in presymptomatic Huntington’s disease. Hum Brain Mapp. 2018;39(10):3871-3883. doi:10.1002/hbm.24217
-
Paulsen JS, Langbehn DR, Stout JC, et al. Detection of Huntington’s disease decades before diagnosis: The Predict-HD study. J Neurol Neurosurg Psychiatry. 2008;79(8):874-880. doi:10.1136/jnnp.2007.128728
-
Zhang Y, Long JD, Mills JA, Warner JH, Lu W, Paulsen JS. Indexing disease progression at study entry with individuals at-risk for Huntington disease. Am J Med Genet Part B Neuropsychiatr Genet. 2011;156(7):751-763. doi:10.1002/ajmg.b.31232
-
Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774-781. doi:10.1016/j.neuroimage.2012.01.021.FreeSurfer
-
Lyu I, Kang H, Woodward ND, Styner MA, Landman BA. Hierarchical spherical deformation for cortical surface registration. Med Image Anal. 2019;57:72-88. doi:10.1016/j.media.2019.06.013
-
Parvathaneni P, Bao S, Nath V, et al. Cortical Surface Parcellation Using Spherical Convolutional Neural Networks. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics). 2019;11766 LNCS:501-509. doi:10.1007/978- 3-030-32248-9_56
-
Klein A, Canton TD, Ghosh SS, et al. Open labels: online feedback for a public resource of manually labeled brain images. 16th Annu Meet Organ Hum Brain Mapping. Published online 2010:84358.
-
Moorhead TWJ, Harris JM, Stanfield AC, et al. Automated computation of the Gyrification Index in prefrontal lobes: Methods and comparison with manual implementation. Neuroimage. 2006;31(4):1560-1566. doi:10.1016/j.neuroimage.2006.02.025
-
Roberts M, Hanaway J, Morest DK. Atlas of the Human Brain in Section. 2nd ed. Lea & Febiger; 1970.
-
Lyu I, Kim SH, Woodward ND, Styner MA, Landman BA. TRACE: A Topological Graph Representation for Automatic Sulcal Curve Extraction. IEEE Trans Med Imaging. 2018;37(7):1653-1663. doi:10.1109/TMI.2017.2787589
-
Alin A. Multicollinearity. Wiley Interdiscip Rev Comput Stat. 2010;2(3):370-374. doi:10.1002/wics.84
-
Verbeke G, Molenberghs G. Linear Mixed Models for Longitudinal Data. Print. Spinger; 2000. https://link.springer.com/chapter/10.1007/978-0-387-22775-7_3
-
Worsley KJ, Andermann M, Koulis T, MacDonald D, Evans AC. Detecting changes in nonisotropic images. Hum Brain Mapp. 1999;8(2-3):98-101. doi:10.1002/(SICI)1097- 0193(1999)8:2/3<98::AID-HBM5>3.0.CO;2-F
-
Taylor JE, Worsley KJ. Detecting sparse signals in random fields, with an application to brain mapping. J Am Stat Assoc. 2007;102(479):913-928. doi:10.1198/016214507000000815
-
Bates D, Mächler M, Bolker BM, Walker SC. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67(1). doi:10.18637/jss.v067.i01
-
Worsley K, Taylor J, Carbonell F, et al. SurfStat: A Matlab toolbox for the statistical analysis of univariate and multivariate surface and volumetric data using linear mixed effects models and random field theory. Neuroimage. 2009;47:S102. doi:10.1016/s1053-8119(09)70882-1
-
Han X, Jovicich J, Salat D, et al. Reliability of MRI-derived measurements of human cerebral cortical thickness: The effects of field strength, scanner upgrade and manufacturer. Neuroimage. 2006;32(1):180-194. doi:10.1016/j.neuroimage.2006.02.051
-
Hong EP, MacDonald ME, Wheeler VC, et al. Huntington’s Disease Pathogenesis: Two Sequential Components. J Huntingtons Dis. 2021;10(1):35-51. doi:10.3233/JHD- 200427
-
Mangin JF, Rivière D, Duchesnay E, et al. Neocortical morphometry in Huntington’s disease: Indication of the coexistence of abnormal neurodevelopmental and neurodegenerative processes. NeuroImage Clin. 2020;26(February):102211. doi:10.1016/j.nicl.2020.102211
-
Scahill RI, Zeun P, Osborne-Crowley K, et al. Biological and clinical characteristics of gene carriers far from predicted onset in the Huntington’s disease Young Adult Study (HD-YAS): a cross-sectional analysis. Lancet Neurol. 2020;19(6):502-512. doi:10.1016/S1474-4422(20)30143-5
-
Nopoulos P, Magnotta VA, Ph D, et al. Morphology of the Cerebral Cortex in Preclinical Huntington’s Disease. Am J Psychiatry. 2012;164(September 2007):1428- 1434.
-
Brown K. Encyclopedia of Language and Linguistics. Vol 1. Elsevier; 2005.